A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes
- PMID: 23160176
- PMCID: PMC3542571
- DOI: 10.1093/gbe/evs100
A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes
Abstract
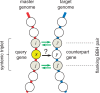
Orthologous relationships between genes are routinely inferred from bidirectional best hits (BBH) in pairwise genome comparisons. However, to our knowledge, it has never been quantitatively demonstrated that orthologs form BBH. To test this "BBH-orthology conjecture," we take advantage of the operon organization of bacterial and archaeal genomes and assume that, when two genes in compared genomes are flanked by two BBH show statistically significant sequence similarity to one another, these genes are bona fide orthologs. Under this assumption, we tested whether middle genes in "syntenic orthologous gene triplets" form BBH. We found that this was the case in more than 95% of the syntenic gene triplets in all genome comparisons. A detailed examination of the exceptions to this pattern, including maximum likelihood phylogenetic tree analysis, showed that some of these deviations involved artifacts of genome annotation, whereas very small fractions represented random assignment of the best hit to one of closely related in-paralogs, paralogous displacement in situ, or even less frequent genuine violations of the BBH-orthology conjecture caused by acceleration of evolution in one of the orthologs. We conclude that, at least in prokaryotes, genes for which independent evidence of orthology is available typically form BBH and, conversely, BBH can serve as a strong indication of gene orthology.
Figures




Similar articles
-
Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals.Genome Biol Evol. 2013;5(10):1800-6. doi: 10.1093/gbe/evt132. Genome Biol Evol. 2013. PMID: 24013106 Free PMC article.
-
Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context.Genome Res. 2001 Mar;11(3):356-72. doi: 10.1101/gr.gr-1619r. Genome Res. 2001. PMID: 11230160
-
Operon conservation from the point of view of Escherichia coli, and inference of functional interdependence of gene products from genome context.In Silico Biol. 2002;2(2):87-95. In Silico Biol. 2002. PMID: 12066843
-
Ancient origin of the tryptophan operon and the dynamics of evolutionary change.Microbiol Mol Biol Rev. 2003 Sep;67(3):303-42, table of contents. doi: 10.1128/MMBR.67.3.303-342.2003. Microbiol Mol Biol Rev. 2003. PMID: 12966138 Free PMC article. Review.
-
Orthologs, paralogs, and evolutionary genomics.Annu Rev Genet. 2005;39:309-38. doi: 10.1146/annurev.genet.39.073003.114725. Annu Rev Genet. 2005. PMID: 16285863 Review.
Cited by
-
Essentiality Is a Strong Determinant of Protein Rates of Evolution during Mutation Accumulation Experiments in Escherichia coli.Genome Biol Evol. 2016 Sep 26;8(9):2914-2927. doi: 10.1093/gbe/evw205. Genome Biol Evol. 2016. PMID: 27566759 Free PMC article.
-
OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy.Genome Biol. 2015 Aug 6;16(1):157. doi: 10.1186/s13059-015-0721-2. Genome Biol. 2015. PMID: 26243257 Free PMC article.
-
Structural and functional analysis of the finished genome of the recently isolated toxic Anabaena sp. WA102.BMC Genomics. 2016 Jun 13;17:457. doi: 10.1186/s12864-016-2738-7. BMC Genomics. 2016. PMID: 27296936 Free PMC article.
-
GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis.Appl Environ Microbiol. 2013 Dec;79(24):7696-701. doi: 10.1128/AEM.02411-13. Epub 2013 Oct 4. Appl Environ Microbiol. 2013. PMID: 24096415 Free PMC article.
-
The genome sequence of 'Mycobacterium massiliense' strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level.PLoS One. 2013 Nov 27;8(11):e81560. doi: 10.1371/journal.pone.0081560. eCollection 2013. PLoS One. 2013. PMID: 24312320 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources


